2022 Director’s Challenge Awards
This cycle, the program supported investigator-initiated, collaborative, and interdisciplinary projects that employ engineering and/or physical science approaches to problems in biology and medicine. The program made six 2-year awards, ranging in amount from $194,000 to $250,000 per year. The currently funded projects, project leaders, and collaborating Institute/Centers are:
- Quantitative Modeling of Viral Propagation Using Robotic/Machine Learning
- Nihal Altan-Bonnet in collaboration with NCI
- Transcriptomic Analysis of Rare Virus-Infected and Tumor Cells by FIND-Seq
- Eli Boritz in collaboration with NCI
- New Nanobody-Based Tools for Improved Tumor Visualization In Vivo
- Ross Cheloha in collaboration with NCI
- Machine Vision-Enabled Behavioral Tracking for Cross-Species Extrapolation
- Dondrae Coble in collaboration with NIBIB and NIDDK
- 4D Map of the Tubulin Code and its Readers in the Human Neuron
- Antonina Roll-Mecak in collaboration with NIBIB
- Point of Care Assay for Detection of Pulmonary Mycobacteria and Lung Injury
- Adrian Marcelo Zelazny in collaboration with NHLBI and Tulane University
Quantitative Modeling of Viral Propagation using Robotic/Machine Learning
Principal Investigator (PI)
Nihal Altan-Bonnet, Ph.D. (NHLBI) In Collaboration with NCI
Project Summary
Viral infection of mammalian hosts’ cells is a multi-step process, involving cellular entry, replication, evasion, viral assembly and egress. This process is highly dynamic with multiple feedbacks on the host cell physiology, e.g., membrane trafficking remodeling, initiation of cellular immune responses, recruitment of egress pathways, cell lysis etc. Such host-pathogen interactions can be critical for viral replication, as demonstrated by previous work by the Altan-Bonnet lab at NHLBI: these interactions, if druggable, offer new opportunities to limit viral replication and propagation. Here we propose to expand the robotic platform developed by the Altan-Bonnet lab at NCI in order to multiplex the timed measurements of viral propagation and associated host cell responses. Ex vivo models of viral replication will be assembled, using varied viruses, host cells and primary immune cells; the dynamics of viral lifecycles and host responses will be quantified using qPCR, cytokine/chemokine bead array and single-cell profiling by spectral cytometry. These high-dimensional datasets will then be analyzed using interpretable machine learning tools, in order to identify the key limiting steps for viral propagation. Model predictions will be further tested and validated in our robotic platform. Such efforts will deliver a quantitative model that identifies key steps in viral replication, accounts for the diversity of host/pathogen interactions, and offers novel drug targets.
Illustration of a robotic arm and SARS-CoV-2 virus.
Transcriptomic Analysis of Rare Virus-Infected and Tumor Cells by FIND-Seq
Principal Investigator (PI)
Eli Boritz, M.D., Ph.D. (NIAID) In Collaboration with NCI
Project Summary
In recent years, emerging microfluidic technologies have enabled single cell sequencing in diverse areas of biomedical science. However, the unbiased “omic” analysis of very rare cells bearing distinctive chromosomal DNA sequences remains beyond the reach of commercial single-cell platforms. In collaboration with academic engineering groups, we have developed a custom microfluidic process termed Focused Interrogation of cells by Nucleic acid Detection and Sequencing (FIND-Seq), which can sort and sequence the transcriptomes of cells harboring latent HIV DNA genomes from samples consisting of only one HIV-infected cell per thousand. Our initial studies using FIND-Seq revealed transcriptomic signatures of HIV-infected memory CD4 T cells that help explain the persistence of these cells in vivo and suggest important avenues for future HIV cure-related research. Here, we propose collaborative efforts among the NIAID Vaccine Research Center, the NHLBI Laboratory of Myeloid Malignancies, and the Trans-NIH Shared Resource on Biomedical Engineering and Physical Science to establish FIND-Seq for intramural studies of HIV-infected cellular reservoirs as well as residual malignant cells responsible for leukemic relapse. These efforts will lead to important insights into HIV persistence and the treatment of leukemia and can also help advance other NIH studies in which defining the biology of rare cells harboring characteristic DNA sequences is critical.
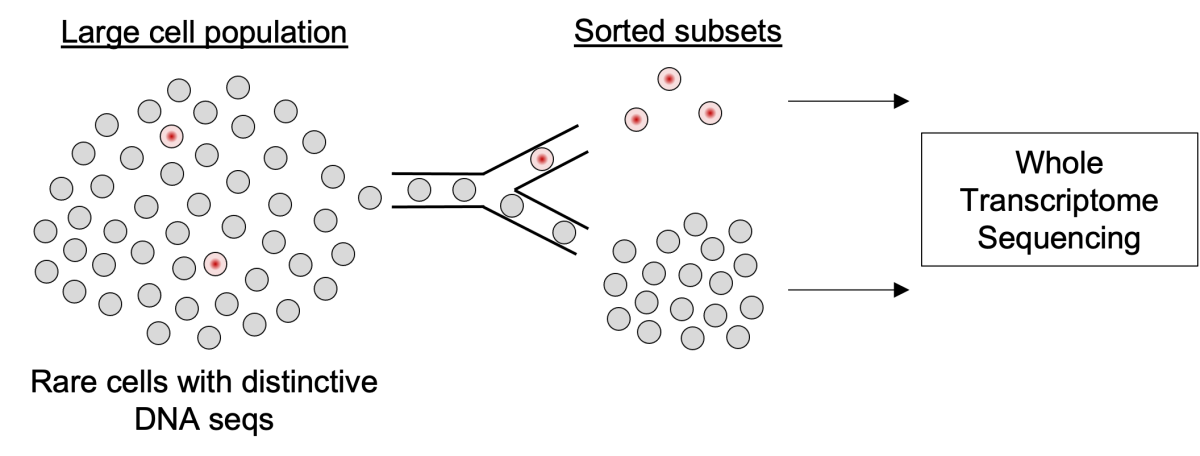
The FIND-Seq process uses droplet microfluidics to capture transcriptomes from millions of single cells, followed by the detection and sorting of droplets based on the presence of specific DNA sequences present in rare cells of interest.
New Nanobody-Based Tools for Improved Tumor Visualization In Vivo
Principal Investigator (PI)
Ross Cheloha, Ph.D. (NIDDK) In Collaboration with NCI
Project Summary
Molecularly targeted imaging agents allow for visualization of tumors not otherwise detectable by conventional methods. Full-length antibodies have been used as imaging agents, however, their long circulating time in vivo is suboptimal for clinical applications. Single domain antibody fragments (nanobodies, Nb) possess several useful properties for imaging. They are easily engineered, non-immunogenic, exhibit a short in vivo half-life, and have been successfully used as positron emission tomography (PET) agents to bind to tumor-associated targets in clinical trials. However, current methods to engineer Nbs as imaging agents are non-specific, adversely affect affinity, and those that are site-specific have poor yields. Using our novel self-labeling, site-specific Nb labeling technology, we hypothesize that we can create Nb-PET agents for use in animal models of human cancers such as liver cancer (aim 1). Furthermore, we hypothesize that we can apply this technology in a two-step pre-targeting approach to enhance tumor-to-tissue contrast (aim 2). If successful, our proposed work will yield specific advances in molecular imaging and open the door to broad biomedical applications in vitro and in vivo.
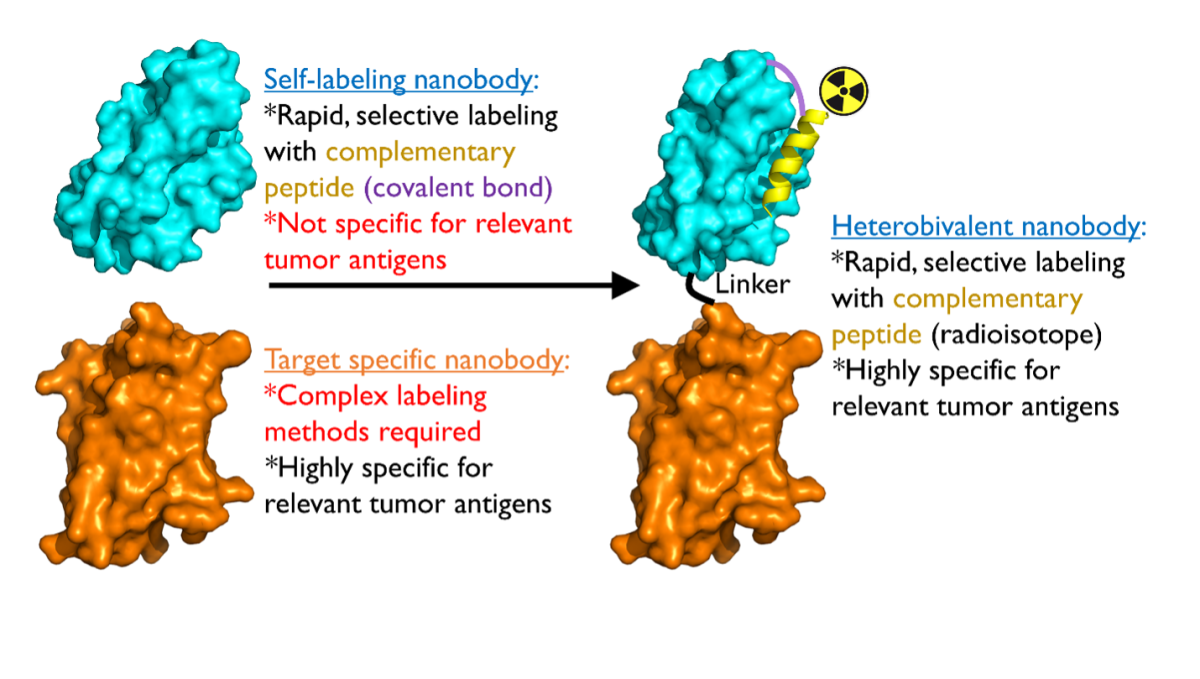
Proposed strategy for using bivalent antibody fragments for visualizing tumors.
Machine Vision-Enabled Behavioral Tracking for Cross-Species Extrapolation
Principal Investigator (PI)
Dondrae Coble, D.V.M. (NIEHS) In Collaboration with NIBIB and NIDDK
Project Summary
Cross-species extrapolation is central to biomedical research where most investigation involves the use of non-human modeling systems as surrogates for human biology. This is particularly relevant in toxicological safety testing where multiple model systems are integrated to make regulatory decisions with profound real-world implications. Synthesis across species leverages evolutionary conservation of adverse outcome pathways (AOP’s) to develop tiered testing pipelines with front-loaded, inexpensive high-throughput screens that advance to more costly labor-intensive screens such as in vivo behavioral testing. Implementation of machine vision-based behavioral decoding in the home environment has the potential to revolutionize this process by providing high-throughput, high-content and continuous behavioral data. Here we propose to combine this approach across two different in vivo models: fruit flies (Drosophila) and mice. This will employ two different hardware systems that we have developed to track these species along with the development of a data analysis pipeline that will be required to process and interpret the data. Drosophila will be used to screen a large set of compounds for more targeted testing in mice. We will build a mechanistic cross-species extrapolation model using both behavioral and molecular (transcriptomic) outcomes. This will ultimately build a broadly applicable cross-species modeling capability with utility far beyond toxicity screens.
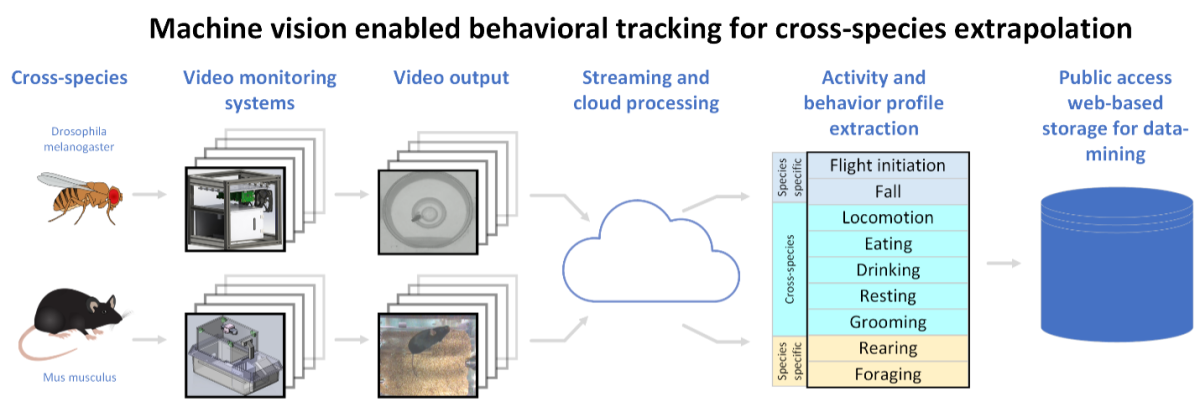
A graphic detailing cross-species extrapolation using two different hardware systems for toxicological testing in the fruit fly and mouse model. Automated video processing algorithms are applied to the video output of the systems. The resulting behavior profiles, along with molecular analysis, are to generate a mechanistic cross-species extrapolation model. This will build a broadly applicable cross-species modeling capability with utility beyond toxicity screens.
4D Map of the Tubulin Code and its Readers in the Human Neuron
Principal Investigator (PI)
Antonina Roll-Mecak, Ph.D. (NINDS) In Collaboration with NIBIB
Project Summary
Life depends on the efficient and organized transport of cellular cargo on microtubules, polymers composed of αβ-tubulins. No cell type is more dependent on this process than the neuron which needs to support fast, efficient communication between compartments that can be more than 1 m apart. As a result, neuronal processes are one of the densest cellular environments, with microtubules spaced as close as 20 nm that support the large continuous volume of bidirectional cellular traffic necessary for survival and optimal function. Neurons have evolved means to organize and differentiate these microtubule tracks by functionalizing tubulin with chemically diverse posttranslational modifications. An emerging paradigm is that these modifications act as a “tubulin code” to regulate microtubule dynamics, mechanics and recruitment of microtubule-associated proteins and microtubule-based motors. The aim of this proposal is to leverage the expertise of our groups on the tubulin code (Roll-Mecak, NINDS) and high-resolution cellular imaging (Hari Shroff, NIBIB and AIM) to generate a complete single-molecule resolution 4D map of the tubulin code and its readers in the human neuron by developing methods for expansion microscopy coupled with single molecule imaging. The methods we develop will be broadly applicable to other structural problems that require nanoscale resolution and molecular contrast.
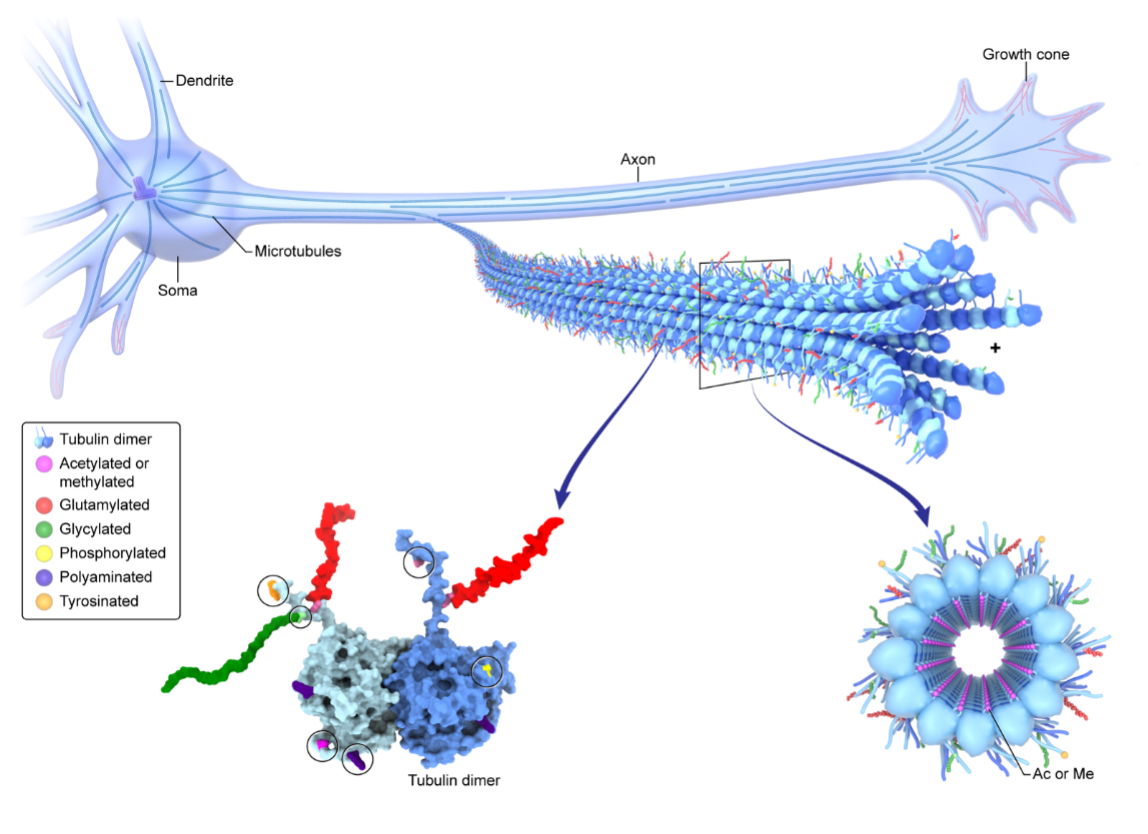
The tubulin code functionalizes the microtubule traffic highways in human neurons.
Point of Care Assay for Detection of Pulmonary Mycobacteria and Lung Injury
Principal Investigator (PI)
Adrian Marcelo Zelazny, Ph.D. (CC) In Collaboration with NHLBI and Tulane University
Project Summary
Mycobacterium species comprise prominent human pathogens causing tuberculosis and leprae, and nontuberculous mycobacteria (NTM). NTM cause lung infections in those with underlying lung disease and some elderly women, soft tissue/surgical site infections, and systemic infections in immunocompromised patients (i.e. AIDS). While tuberculosis rates have been declining in developed nations, NTM disease continues to increase. Diagnostic tools have focused on tuberculosis and are lacking for NTM. An ultrasensitive CRISPR-based assay for detection of circulating M. tuberculosis cell-free DNA (cfDNA) was developed by Dr. Tony Hu and team (longterm NIH collaborators) and showed excellent performance in pediatric and adult TB and HIV-associated TB, 94.1-100% sensitivity. This CRISPR based-assay can be used as cellphone-based point of care assay. Concurrently, novel methods to detect lung and other tissue-specific injury sampling plasma cfDNA have been developed at the NIH. One approach targets tissue-specific DNA methylation signatures of cfDNA as markers of injury. In COVID-19, an increase in lung-specific cfDNA correlated with disease severity. Another approach utilizes chromatin immunoprecipitation sequencing (chipseq) to assess tissue-specific cfDNA. We aim to develop a point of care assay of circulating cfDNA of 1) Most common NTM (M. avium complex, M. kansasii, M. abscessus) and 2) Lung damage through lung-specific DNA methylation and/or promoter chipseq of lung-specific genes.
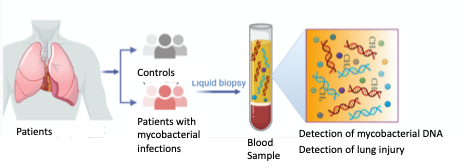
Point of care blood assay for detection of pulmonary mycobacterial infection and lung injury.
This page was last updated on Tuesday, August 2, 2022